The colsample_bylevel parameter in XGBoost controls the subsample ratio of features (columns) for each level of the tree.
It determines the fraction of features that will be randomly sampled at each split, allowing for different subsets of features to be considered at different levels of the tree.
This example demonstrates how to tune the colsample_bylevel hyperparameter using grid search with cross-validation to find the optimal value that balances feature representation at each level of the tree.
import xgboost as xgb
import numpy as np
from sklearn.datasets import make_classification
from sklearn.model_selection import GridSearchCV, StratifiedKFold
from sklearn.metrics import accuracy_score
# Create a synthetic dataset
X, y = make_classification(n_samples=1000, n_classes=2, n_features=20, n_informative=10, random_state=42)
# Configure cross-validation
cv = StratifiedKFold(n_splits=5, shuffle=True, random_state=42)
# Define hyperparameter grid
param_grid = {
'colsample_bylevel': np.arange(0.2, 0.9, 0.1)
}
# Set up XGBoost classifier
model = xgb.XGBClassifier(n_estimators=100, learning_rate=0.1, random_state=42)
# Perform grid search
grid_search = GridSearchCV(estimator=model, param_grid=param_grid, cv=cv, scoring='accuracy', n_jobs=-1, verbose=1)
grid_search.fit(X, y)
# Get results
print(f"Best colsample_bylevel: {grid_search.best_params_['colsample_bylevel']}")
print(f"Best CV accuracy: {grid_search.best_score_:.4f}")
# Plot colsample_bylevel vs. accuracy
import matplotlib.pyplot as plt
results = grid_search.cv_results_
plt.figure(figsize=(10, 6))
plt.plot(param_grid['colsample_bylevel'], results['mean_test_score'], marker='o', linestyle='-', color='b')
plt.fill_between(param_grid['colsample_bylevel'], results['mean_test_score'] - results['std_test_score'],
results['mean_test_score'] + results['std_test_score'], alpha=0.1, color='b')
plt.title('Colsample Bylevel vs. Accuracy')
plt.xlabel('Colsample Bylevel')
plt.ylabel('CV Average Accuracy')
plt.grid(True)
plt.show()
# Train a final model with the best colsample_bylevel value
best_colsample_bylevel = grid_search.best_params_['colsample_bylevel']
final_model = xgb.XGBClassifier(n_estimators=100, learning_rate=0.1, colsample_bylevel=best_colsample_bylevel, random_state=42)
final_model.fit(X, y)
The resulting plot may look as follows:
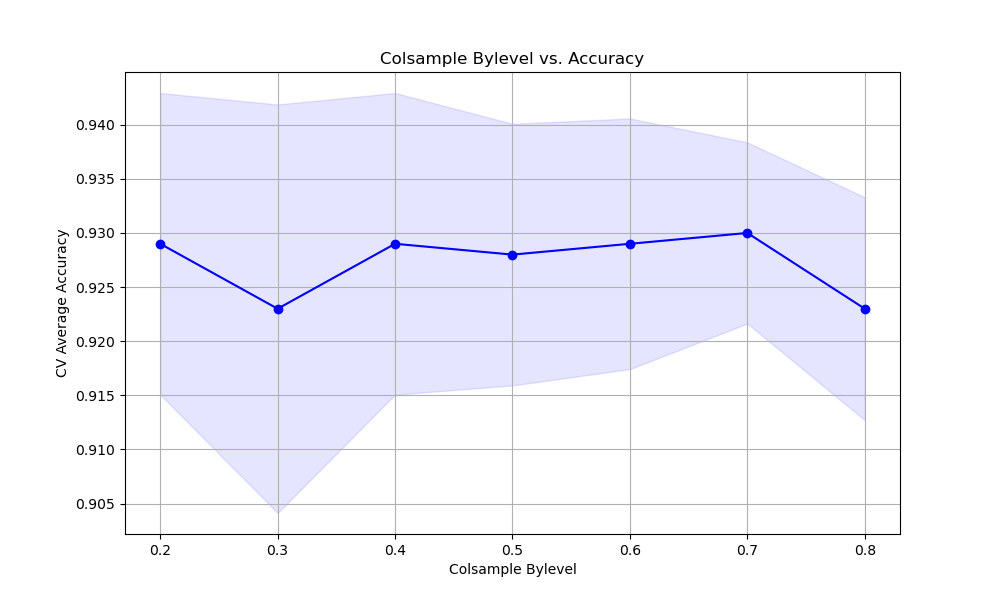
In this example, we create a synthetic binary classification dataset using scikit-learn’s make_classification function. We then set up a StratifiedKFold cross-validation object to ensure that the class distribution is preserved in each fold.
We define a hyperparameter grid param_grid that specifies the range of colsample_bylevel values we want to test, from 0.2 to 0.8 with a step of 0.1.
We create an instance of the XGBClassifier with basic hyperparameters set. We then perform the grid search using GridSearchCV, providing the model, parameter grid, cross-validation object, scoring metric (accuracy), and the number of CPU cores to use for parallel computation.
After fitting the grid search object, we can access the best colsample_bylevel value and the corresponding best cross-validation accuracy using grid_search.best_params_ and grid_search.best_score_, respectively.
We plot the relationship between the colsample_bylevel values and the cross-validation average accuracy scores using matplotlib. We retrieve the results from grid_search.cv_results_ and plot the mean accuracy scores along with the standard deviation as error bars. This visualization helps us understand how the choice of colsample_bylevel affects the model’s performance.
Finally, we train a final model using the best colsample_bylevel value found during the grid search. This model can be used for making predictions on new data.
By tuning the colsample_bylevel hyperparameter, we can find the optimal value that balances feature representation at each level of the tree, allowing the model to consider different subsets of features at different levels and potentially improve its performance.
